Short essay (300 words):
Briefly comment on bioinformatics web app design and usage, including important key considerations, information extraction, typical problems, future challenges, promising directions, and other issues that catch your attention. (20 points)
- Key consideration
- Information extraction
- Typical problems
- Future challenges
- Promising direction
Web skills, one of the significant tools in demonstrating experiment results, have gained increasing importance in bioinformatics students and researchers. Generally, a bioinformatics web application could process some kind of biological data from users’ input and output the annotated or elaborated results, for instance, RNA-seq, GO analysis. In other words, a bioinformatics web application contains three steps, input, processing, and output. Therefore, UI design scripts and data processing scripts would be necessary for bioinformatics people. Regarding the UI design, HTML, CSS and JavaScript would be the top choice, meanwhile, regarding the data processing language, R or Python would be widely used.
Owing to the fact that most bioinformatics people do not possess enough programing knowledge to learn all the language suggested previously, they generally mastered one language, for instance, R. As a result, “shiny” package based on R was devised and researchers could now design and publish their own web application with a few R codes. To be more specific, shiny is a highly assembled R package and sufficient to cover basic requirements for demonstration. Hence, there is no need for bioinformatics researchers to learn extra web design language and let them focus on the data processing part, saving their energy and time.
However, the shiny-based web application has its own drawbacks. Based on my personal experience in this coursework, I found that the tools were limited and could only satisfy the basic layout, rather than fancy, well-designed web pages. Also, since shiny is based on R, researchers’ choice could be restricted to R, but some heated techniques like deep learning could not achieve their full potential on the R platform, so the application area is in a small scope. In the meantime, users have to devote some time to learning the document of shiny, but the overall investment could already support you to learn the normal web designing language HTML, CSS and Javascript. The previously stated reasons could probably account for the limited use of shiny in actual web development in the bioinformatic field, they mostly prefer the normal web development tool. In reality, it is a great way to start by shiny and cultivate some interests in web design and turn to other more general, widely used tools
The layout of the website
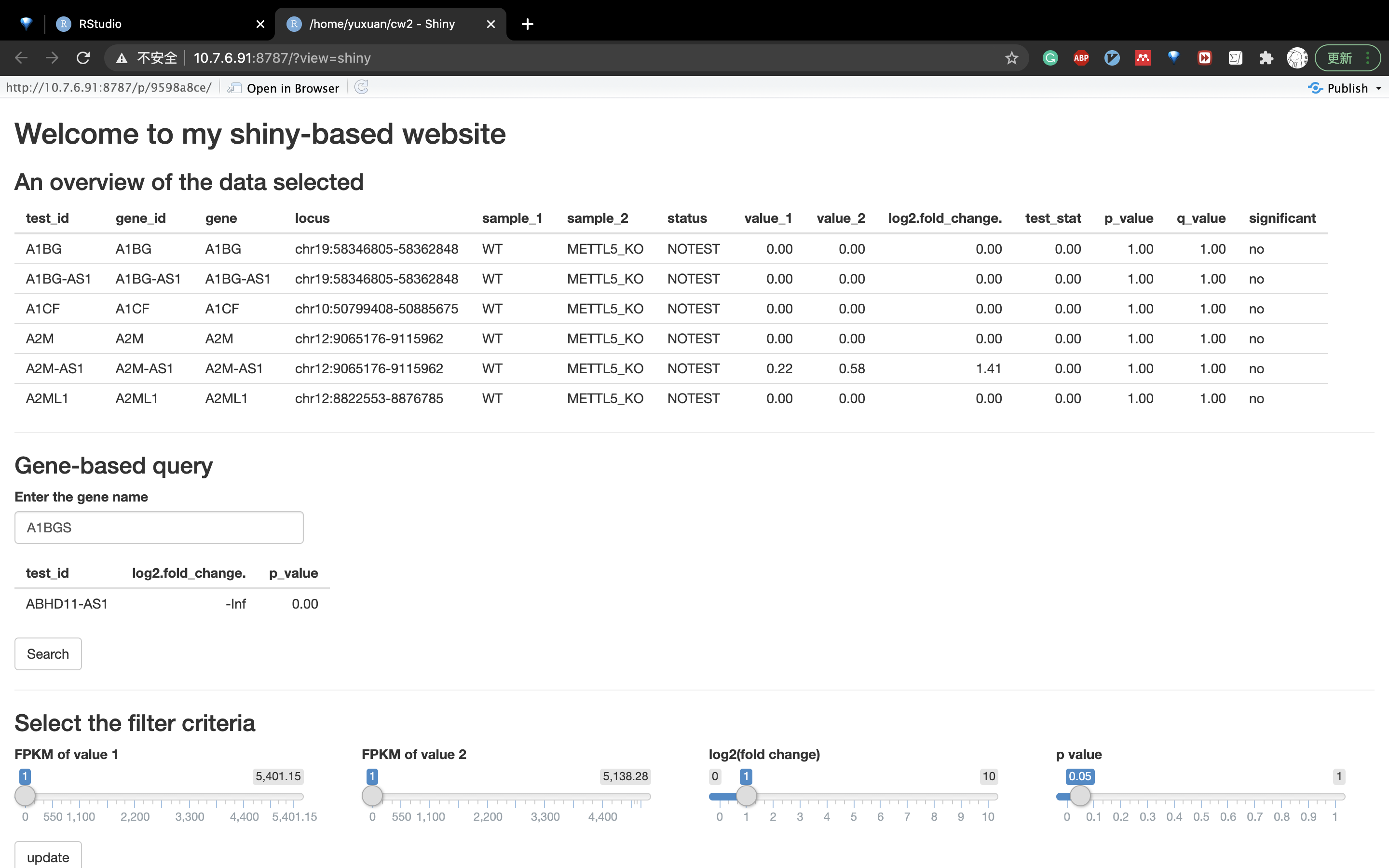
Filtering criteria

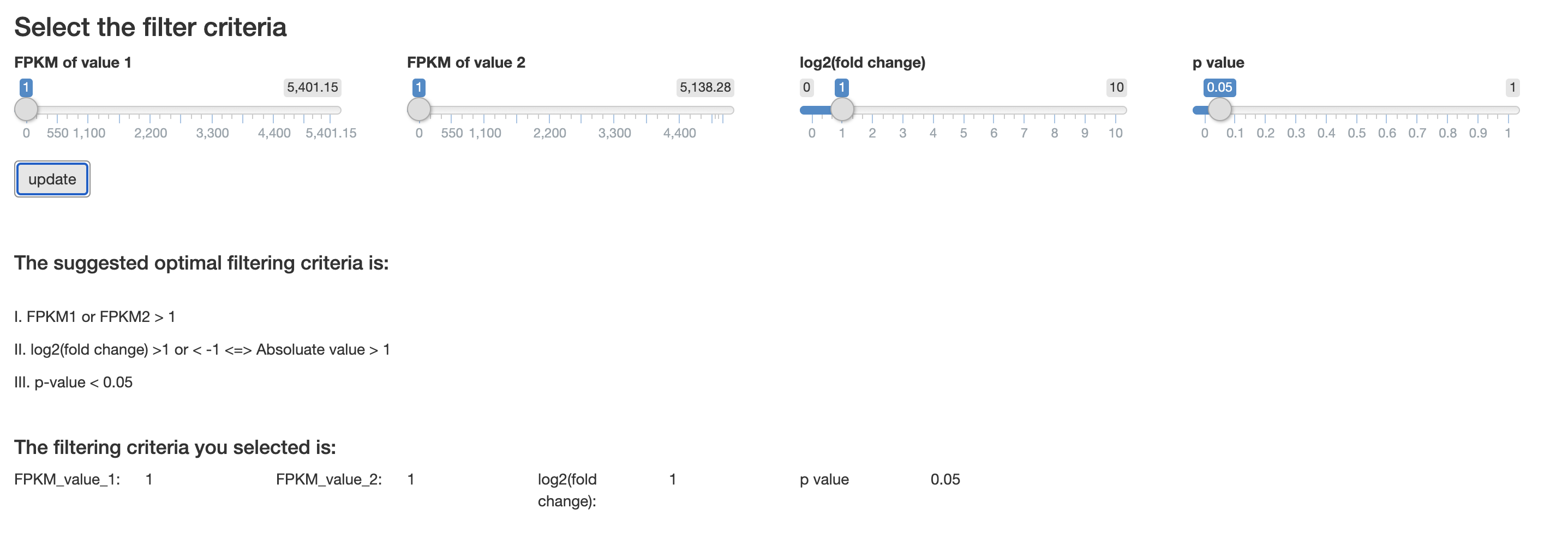
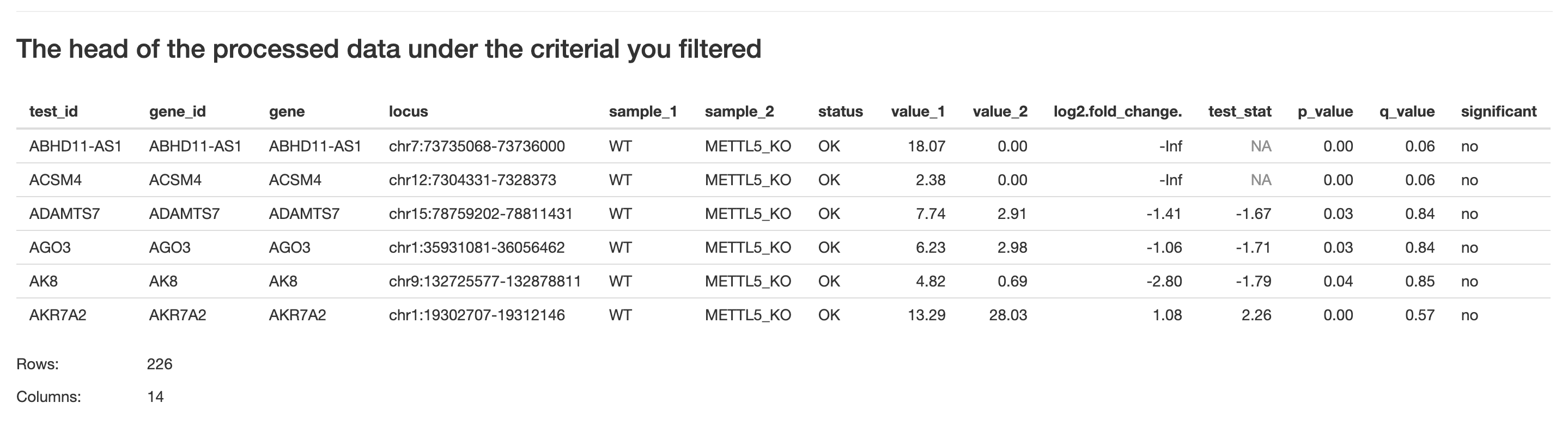

Server
1 | # Title : TODO |
UI
1 | # Title : TODO |
- Post title:Shiny 网页设计
- Post author:Yuxuan Wu
- Create time:2021-04-17 02:24:26
- Post link:yuxuanwu17.github.io2021/04/17/2021-04-17-Shiny-网页设计/
- Copyright Notice:All articles in this blog are licensed under BY-NC-SA unless stating additionally.