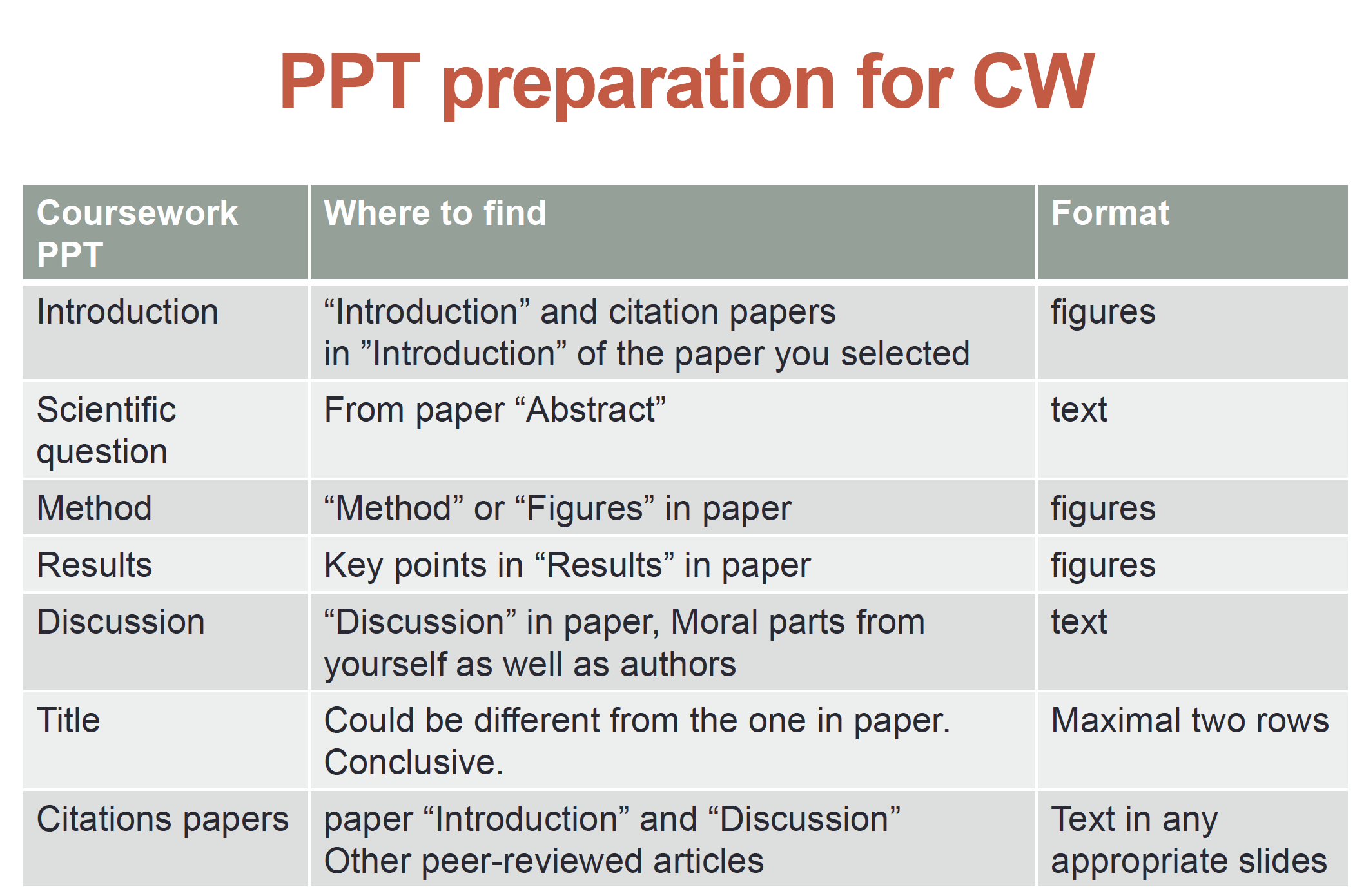
- Give an introduction to enable the audience to follow the rest of your presentation
- Explain the purpose (scientific question) of the research in the paper.
- Briefly highlight one or two key approaches.
- Explain the key findings of the paper
- Discuss the ethical issues involved
==Maximum 20 pages==
Find and digest information about a biotechnological/ ethical issue
Form an opinion on the biotechnological / ethical issue and be able to provide arguments to defend your stance
Explain the technology and illustrate your opinion in a 10’ presentation
Introduction
Genetic diseases of blood cells are prime candidates for treatment through ex vivo gene editing of CD34+ hematopoietic stem/progenitor cells (HSPCs)
Mechanisms:
SCD is caused by a single-nucleotide polymorphism (SNP) in the seventh codon of the gene for
Existing treatment
RBDs are produced from the bone marrow and hematopoietic stem cell
RBCs are produced from repopulating hematopoietic stem cells (HSCs) in the bone marrow (BM), and allogeneic hematopoietic cell transplantation (HCT) from an unaffected human lymphocyte antigen (HLA).matched donor is currently the only lasting cure for SCD
Problems:
- Difficulty in identifying donors
- Toxicity of the transplant regimen
- Graft-versus-host disease
What they did
hematopoietic stem/progenitor cells (HSPCs)
We optimize design and delivery parameters of a ribonucleo protein (RNP) complex comprising Cas9 protein and unmodified single guide RNA, together with a single-stranded DNA oligonucleotide donor (ssODN), to enable efficient replacement of the SCD mutation in human HSPC
ex vivo treated human HSPCs maintain SCD gene edits throughout ==16 weeks== at a level likely to have clinical benefit.
During gene editing, a targeted nuclease creates a double-strand break (DSB) that can be repaired by one of two mechanisms:
error-prone non-homologous end joining (NHEJ) that results in ==genomic insertions and deletions (indels)== not specific.
==templated homology-directed repair== (HDR) to precisely insert, delete, or replace a genomic sequence
The recent development of CRISPR-Cas9, a programmable RNA-guided DNA endonuclease, has ignited an explosion of interest in gene editing to cure many genetic disorders, including SCD.Guided by a single guide RNA (sgRNA), the Cas9 nuclease can be programmed to cut a target locus within the genome, allowing rapid iteration and optimization not possible with other gene editing approaches
Pipeline
an erythroleukemia cell line to explore a panel of Cas9 RNPs that cut near the SCD mutation
RNPs to develop ex vivo editing methods in human HSPCs, achieving up to 33% sequence replacement
We demonstrate efficient correction of the sickle mutation in SCD HSPCs, with corresponding production of WT adult hemoglobin (HbA) RNA and protein in edited, differentiated erythroblasts.
After the edited human HSPCs are engrafted in immunocompromised mice, sequence replacement at the SCD locus is retained 4 months after engraftment at levels likely to have clinical benefit.
Results
- We chose to use inexpensive, commercially available ssODNs and co-delivered these with Cas9 RNP by electroporation. Under this condition, quickly iterate combinations of sgRNA, HDR donor, and editing conditions.
- Two key restrictions when designing sgRNAs and ssODNs for HDR experiments:
- the distance between the sgRNA recognition site and the mutation
- the ability to silently ablate the sgRNA protospacer adjacent motif (PAM).
- ensures that Cas9 cannot recut corrected alleles, thus preventing the introduction of indels into the corrected allele
Find the most active sgRNAs compatible with SCD editing
we identified targets in the first exon of theHBB gene and tested six of them for which the PAM could be silently mutated (G3, G5, G10, G11,G17, and G18) and one for which it could not (G7) (Fig. 1A)
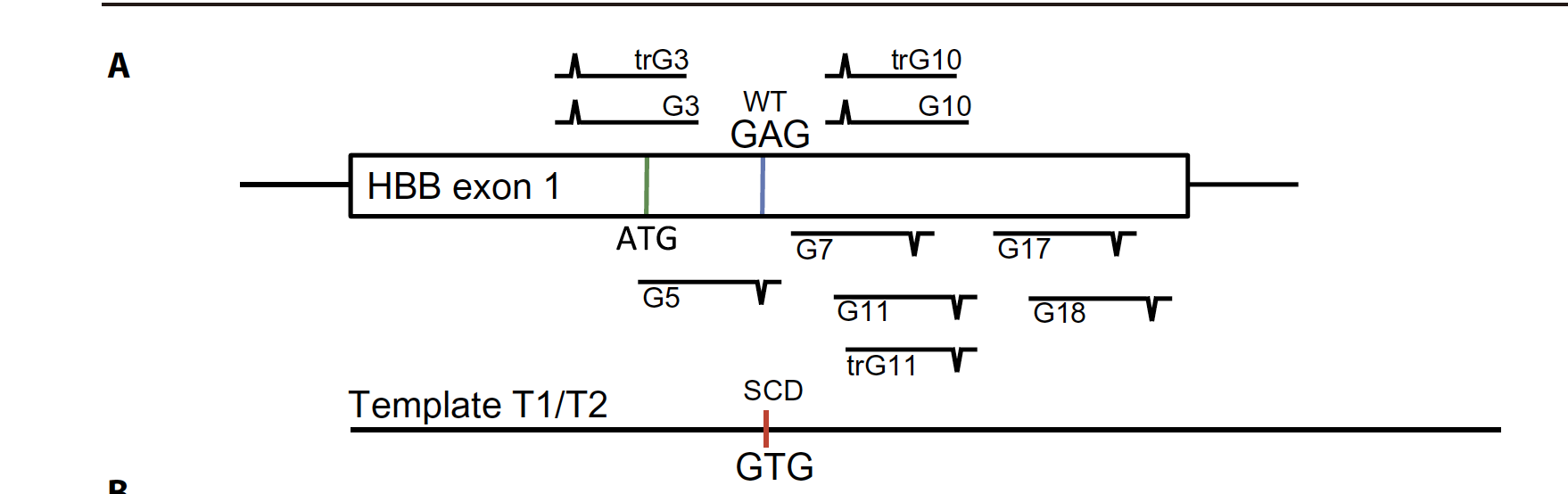
Editing the SCDSNP in K562 cells. (A) Schematic depicting the experimental approach to editing in K562 cells. A panel of 10 sgRNAs that cut within 100 base pairs (bp) of the SCD SNP was selected. A WT-to-SCD edit was programmed by an ssDNA template (T1) bearing silent PAM mutations for the sgRNAs, which also introduces an sfc I restriction site.
T7 endonuclease 1 assay depicting indel formation in pools of cells edited by candidate RNPs.
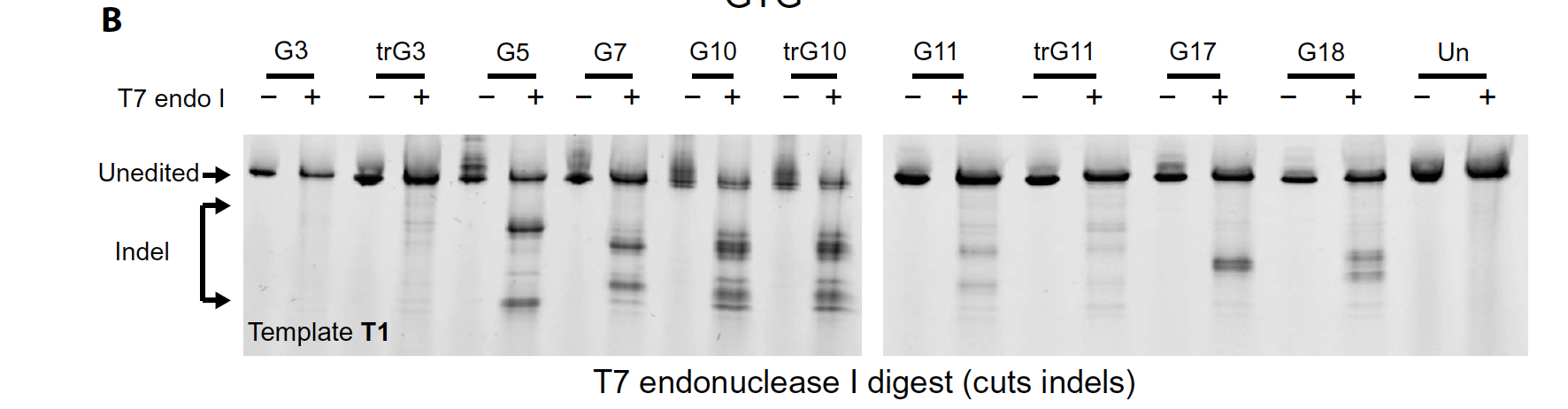
(C) Editing of candidate sgRNAs detected by Sfc I digestion. G5, G10, and a truncated variant, trG10, efficiently edit in K562 cells.
(D) Gene modification of select sgRNAs and templates at the SCD SNP, assessed by NGS. See fig. S1 for definitions of donors T1 and T2. Data are the average of three biological replicates, and error bars indicate SD.
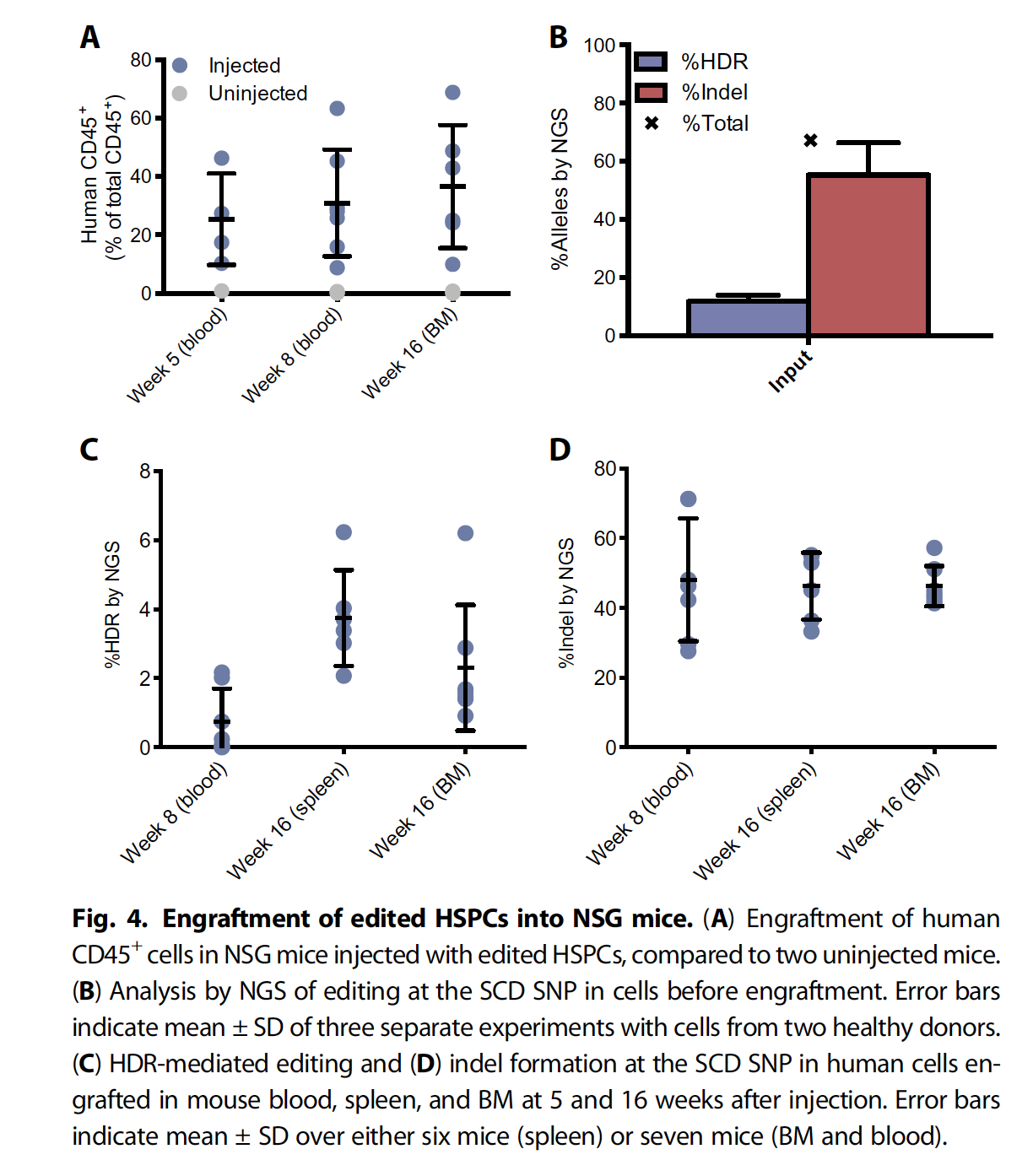
FigA: (CD45+) Substantial numbers of hematopoietic cells were detectable in BM of all injected mice at 16 weeks
Genotyping of edited HSPCs was performed by NGS immediately after editing and before injection (Fig. 4B) and from mice at weeks 8 (blood) and 16 (BM and spleen脾脏) (Fig. 4, C and D). The input populations had 55 ± 19% indel alleles and 11.8 ± 3.7%HDR alleles (mean ± SD) (Fig. 4B).
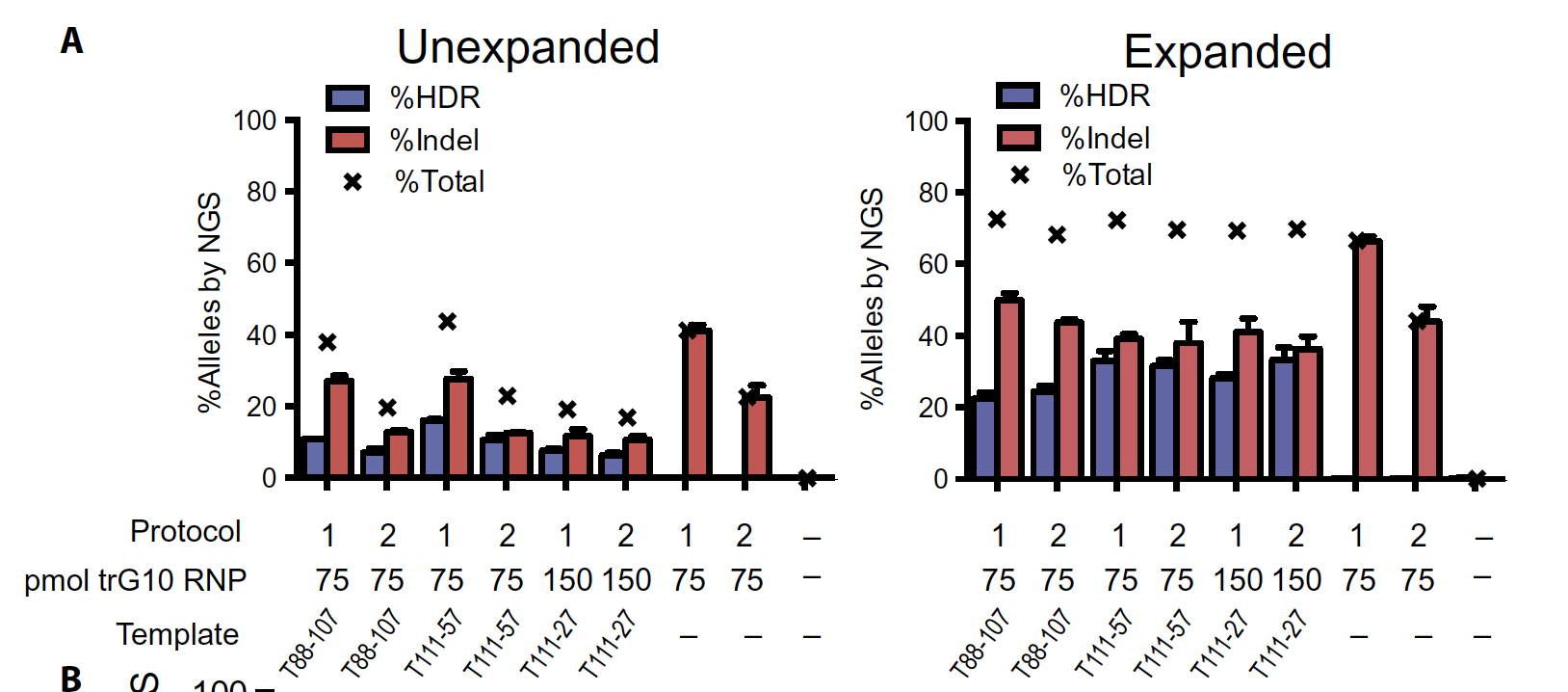
To selectively interrogate editing in viable cells, we used NGS to assay HSPCs cultured under erythroid expansion conditions for 7 days after editing (erythroidexpanded), along with edited HSPCs cultured for only 2 days (unexpanded).
We observed appreciable initial levels of editing at the sickle SNP inHSPCs, with HDR rates between 6 and 11% (Fig. 2, A and B). After 5 days of erythroid expansion, HDR rates increased, with up to 33% editing at both high and low doses of RNP (150 and 75 pmol, respectively)
Total editing (%HDR + %NHEJ) in expanded HSPCs was between 66 and 72%, indicating good delivery of the trG10 RNP to HSPCs. In general, higher editing was accompanied by some reduction in viability, as measured by fewer cells remaining after treatment, particularly at a high, 150 pmol dose of RNP
NGS analysis revealed consistently high levels of indel alleles after 16 weeks
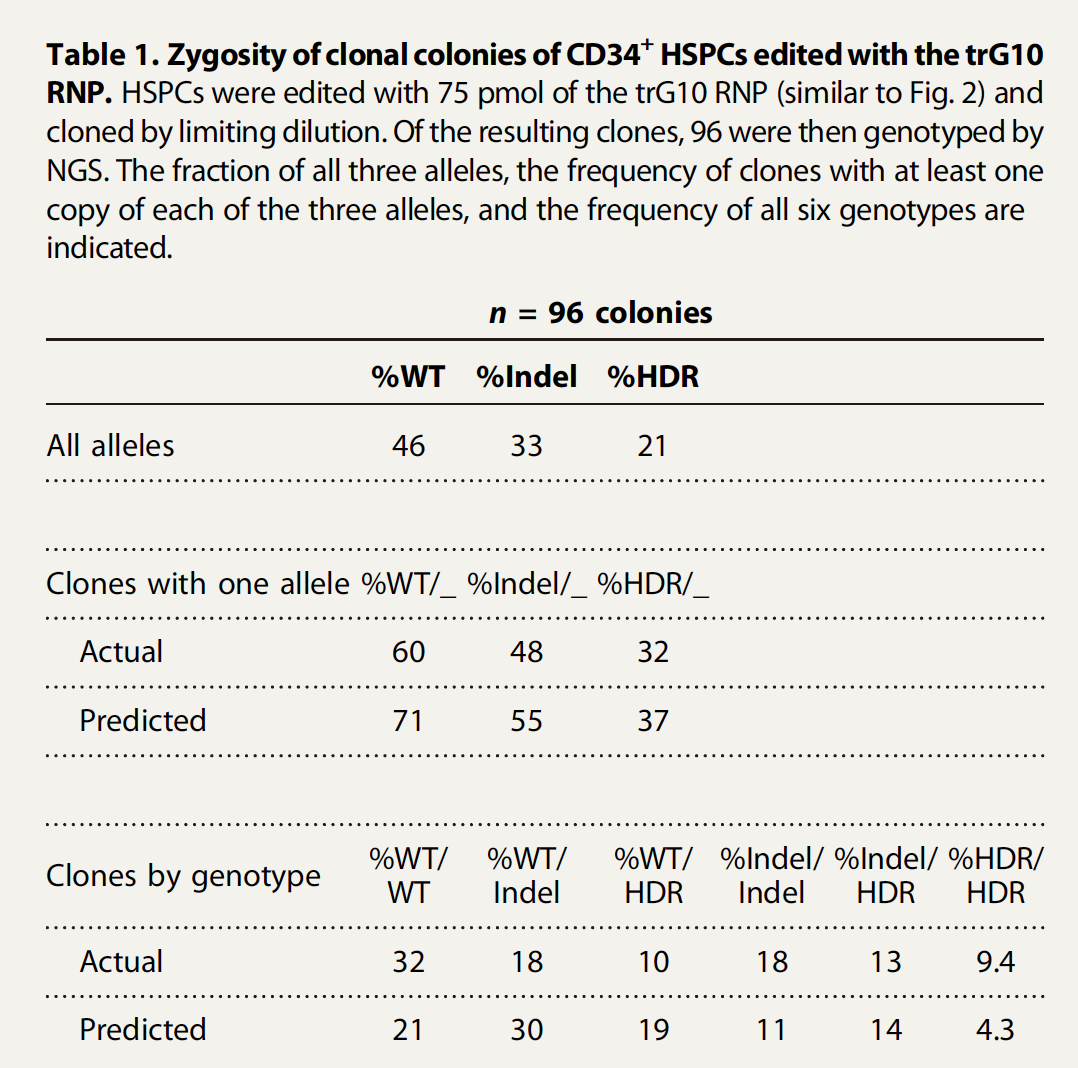
After 14 days of growth, 96 edited clones were individually genotyped by multiplexed NGS (Table 1). In this experiment, 21% of alleles were HDR. However, these alleles were spread among 32% of the cells. This increased prevalence of edited cells relative to edited alleles is predicted by the independent assortment of alleles within a population (although we observed that homozygous genotypeswere still overrepresented relative to a prediction based on random assortment)
In this model, edited human stem cells must engraft in the mouse and persist over several months for them to be observed in subsequent experiments
After 16weeks, progenitor cells should be lost from the system, and human cells in the BM should be derived from long-term repopulating stem cells within the initial HSPC population
Discussion
- Unlike gene therapy using integrating viral vectors, wherein regulation of the introduced gene may be compromised and endogenous genes may be disrupted, gene editing corrects the disease mutation at the endogenous locus
- No need to introduce a nonnative selection marker
- edits do not typically confer a selective advantage in the target cell compartment, as is the case for correction of the HBB sickle mutation in BM stem cells
We used K562 cells to iterate combinations of Cas9 RNPs and ssODNs that edit the HBB gene and then applied these reagents to human HSPCs.
These reagents efficiently induce HDR-mediated editing of the SCD mutation in HSPCs with minimal genic off-target activity.
we found no apparent decrease in off-target activity compared to the full-length guide, possibly due to the extremely high homology between the on- and off-target sites. RNP delivery has been associated with reduced off-target effects
this modality could allow efficient editing of disease mutations in HSPCs using full-length unmodified sgRNAs such as G10
To reduce off-target activity, we tested several recently characterized high-fidelity Cas9 variants as RNPs, but chose to pursue WT Cas9 due to their reduced on-target activity
the co-delivered RNP/ssODN approach described here is nonviral, modular, and readily optimized
However, HDR was still diminished relative to the input CD34+ populations. Reductions in HDR frequency during long-term engraftment have been observed previously and remain a major impediment to bringingHDR-based therapies to the clinic (14, 15, 21). It may be thatHDR-editedHSCs do not engraft as well as unedited cells, that theHDRdonors are themselves toxic, or that HSCs are intrinsically more difficult to edit than other CD34+ cells
Corrected SCD HSPCs produced greatly reduced sickle and increased WT hemoglobin protein and b-globin mRNA
Limitation
- it is possible that the allelic correction frequency we observed in the HSPC population will ameliorate but not eliminate the clinical expression of SCD.
- it is still an intensive procedure, requiring myeloablative conditioning, which can have serious side effects.
- clinical translation of an HSCT-based therapy is not feasible in developing countries, where SCD is most prevalent. We anticipate that further developments will address these remaining issues
Ethical problems
- But a capability for making precise changes to the human genome raises all kinds of difficult questions about how far we should go with it
- Should genome engineering be restricted to the avoidance of genetic disease, or might it be justified for genetic enhancement
- How can we distinguish one from the other
- Where are the limits on the possible or permissible – giving us infrared vision, say, or tolerance to extreme cold, or the ability to photosynthesise?
- Should we use Crispr on the human germline, so that modifications are inherited by future generations?
The last issue has become particularly explosive since the revelation that Crispr was used in 2018 by Chinese biologist He Jiankui to modify twin embryos used for IVF, resulting in the birth of two girls allegedly containing alleles that would confer protection from infection by HIV. It wasn’t just that He bypassed ethical regulations, nor even that he chose to use germline editing for pre-emptive protection. It was also that he did the job rather poorly and without any clear evidence that the procedure was safe.
inheritable modifications
it is not necessarily as accurate as it is sometimes portrayed
lead to off-target modifications, the health consequences of which are unknown and unpredictable
worldwide legislation should be prepared
CRISPR-Cas9 should be allowed for use in the creation of human disease models, and in understanding the development and molecular mechanisms of diseases
Should be prohibitive for the purposes of eugenics or enhancement.
Heredity problems
presentation script
Hello everyone, my name is yuxuan wu and thank you for the time in listening my presentation. Today my topic is about genome editing by Crisper technique, specifically the sickle cell mutation treatment in human adult hematopoietic stem cells.
Here is a brief outline of my whole presentation. I would firstly give you some background information about this diseases over the world so you could easily catch up. Then I would analyze the mechanisms about the sickle cell diseases and the newly raised heated technique Crispr , for instance, how they originated and how they worked. Then I would focus on the paper I selected to let you guys know some advancement in this areas. Additionally, since the crispr technique involves the edit of genetic materials, which means some ethical problems might raised and I would discuss this point later
Now we move on the first part, introduction part. As can be seen from here that the sickle cell diseases is a group of blood disorders typically inherited from a person’s parents, which means it is closely related to heredity. To be more specific, the mutation occurs in chromosome 11 that results in an abnormality in the oxygen-carrying protein hemoglobin found in the red blood cell. The abnormal cell would feature in a sickle-like shape. Generally speaking, the disease would typically begin around 5 to 6 month which could results severe symptoms like early death, bacterial infection and stroke. The average life expectancy in the developed world is 40 to 60 years old. However, for people in developing world, where most cases occurred, that would lead to an early death, or infant death.
2min
Here I would introduce the mechanisms behind the SCD. As I mentioned before, this is a genetic diseases caused by the base substitution, leading to production of abnormal hemoglobin S. As a result, the capacity for carrying oxygen would be decreased significantly, causing the vaso-occlusion and the severe symptoms.
This slide is a more vivid example with clear annotation. Figure A shows the normal red blood cells passing the vessel. We could see that the normal cells could pass the cross-section without any difficulty while in sickled red blood cells, they would block the vessels and cause severe negative effects.
In order to cure this diseases, novel approaches like crispr technique started to play a significant role. This technique is a gene editing approach to enable efficient replacement of the SCD mutation. Here, the abnormal cells would have a markedly shorter life span in circulation compared to the wild type, which means the low levels of genotypic correction are predicted to generate a clinical benefit.
4 min
Now, we finally moved to the paper part to present a novel method to cure the disease. Here I demonstrate the whole pipeline. They firstly design a programmable RNA-guided DNA endonuclease and edit the target cells ex vivo. One of the key features in crispr was its precision, the designed RNA-guided DNA would find the SCD mutation site and correct it. The corrected cell would cultivate to erythroblasts [ɪˈrɪθrə(ʊ)blæst], which is a stage in the development of the red blood cells. The next step would be the comparison between the wild type to check its correction efficiency. Once checked, engraft the samples in the immunocompromised mice for four months. After four months, the mice would sacrifice to check the efficacy of this technique.
Here is the results and discussion part. These are the two figures that compare the cell condition under two days cultivation and seven days cultivation. Two terminologies were extremely important here, HDR and Indels, both representing the modification in the sequence, but HDR rate is the precisely modification while Indels are not. We could observe that the initial HDR level would be around 6% to 11%, after 5 days of cultivation, HDR rates increased, with up to 33% editing under different conditions. Total editing (%HDR + %NHEJ) in 7 days cultivation was around 70%, indicating good delivery of ribonucleous proteins.
6min (5:30min)
Now we could engraft the successfully cultivated cells in the immunocomprised mice. The paper made great efforts to ensure the low off-target activities, but considering the time limit, I therefore would not include this part. Here I would explain some abbreviations that HSPC was stand for the hematopoietic stem/progenitor cells (HSPCs). From figure A, it is clear that the mice injected with modified HSPC could have a higher percentage compared to those uninjected mice.Therefore, the figure suggested that the mice injected hematopoietic stem cells with corrected SCD mutation produced greatly reduced sickle and increased wild type, normal hemoglobin. Furthermore, the Next generation sequence techniques revealed the consistently high levels of indel alleles even after 16 weeks, suggesting possible clinical benefits.
Several limitation could still occurred in this experiment. Firstly, the allelic correction frequency would ameliorate, or in other words, the performance would diminish and could not last permanently. Secondly, the crispr technique is still an intensive procedure, which can have unknown side effects. Thirdly, the clinical translation of HSCT-based therapy is not feasible in most developing countries, while these places are SCD most prevalent. ==(Hematopoietic stem cell transplantation (HSCT) )==
7:30
Next part would be ethical problems related to this technique, especially the treatment including the modification of genetic materials. Several questions need to be answered if we would like to step further. For instance, how far should we go with it? Should genome engineering be restricted to the avoidance of genetic disease? Might it be justified for genetic enhancement? Should we use Crispr on the human germline, so that modifications are inherited by future generations?
But at the time we could not even answer the previous problems, He jiankui conducted an astonishing experiments. He modified the twin embryos used for the protected from infection by HIV, but the safety of this experiment could not be ensured. Additionally, the high potential of the off-target modification made these twins suffer from unknown and unpredictable health consequences. Furthermore, we got no ideas about whether the modification would be inheritable or not.
Here is my personal view about the ethical problems. I divided into two sections, either is applicable or prohibitive. We should publish restricted worldwide legislation to ensure the monitor of experiments which could have high potential of unethical problems. We could allow the creation of human diseases models and use the technique to elucidate the molecular mechanisms of diseases. These three points are applicable and the following three points should be prohibited strictly. For instance, the breeding of the human species, the germ-line editing in heredity non target genetic mutations. And the last point is definitely not allowed, that is to create weapons, that will cause disasters to human beings all over the world
This is the reference I used in this presentation. That’s all for my whole presentation, thank you for listening.
- Post title:biotechnology presentation
- Post author:Yuxuan Wu
- Create time:2021-03-10 05:50:03
- Post link:yuxuanwu17.github.io2021/03/10/2021-03-10-BIO304-biotechnology-presentation/
- Copyright Notice:All articles in this blog are licensed under BY-NC-SA unless stating additionally.